- 翰林提供学术活动、国际课程、科研项目一站式留学背景提升服务!
- 400 888 0080
IB DP Biology: SL复习笔记3.1.4 Genome
Genome
- The total of all the genetic information in an organism is called the genome of the organism
- This is a complete set of genes present within every cell of an organism
- This includes all genes as well as non-coding DNA sequences
- Mitochondrial DNA and chloroplast DNA are included in the genome
- In a prokaryote cell, plasmid DNA is included in its genome
Comparing Genome Size
- Advances in technology have allowed scientists to write the whole sequence of the genes within an organism's genome
- Genome-wide comparisons can now be made between individuals and between species
- Sequencing projects have read the genomes of a wide range of organisms from bacteria to humans
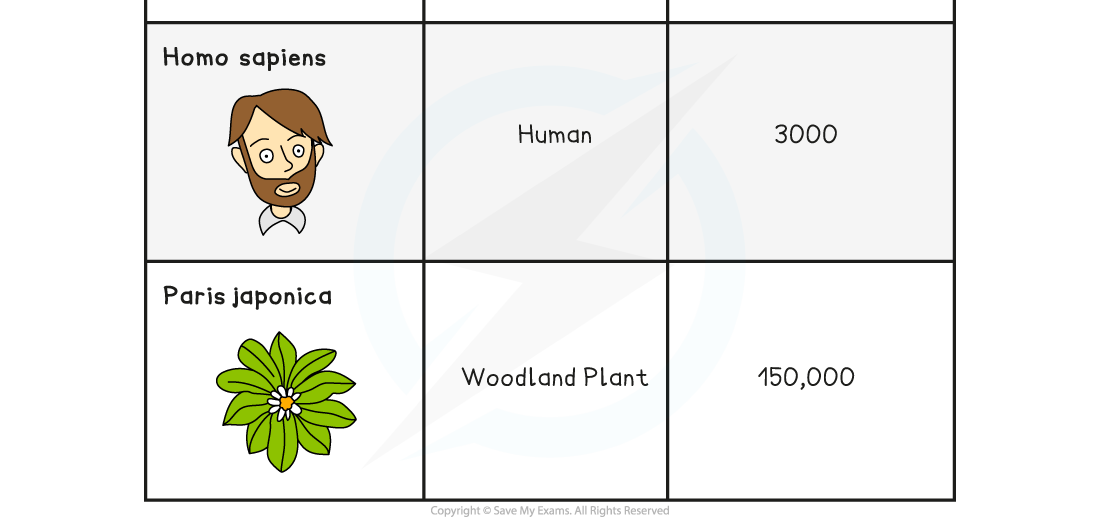
- Genome sizes can differ in different organisms:
- Viruses and bacteria tend to have very small genomes
- Prokaryotes tend to have smaller genomes than eukaryotes
- The size of plant genomes can vary widely
Comparing the Genome Size of Different Organisms Table

Human Genome Project
- The Human Genome Project (HGP) was an international, collaborative, research programme to sequence the entire human genome
- The work began in 1990, was publicly funded, and shared among more than 200 laboratories around the world, avoiding duplication of effort
- Different labs sequenced different chromosomes
- DNA samples were taken from multiple people around the world, sequenced, and used to create a reference genome
- Because of rapid improvements in base sequencing technology the project finished ahead of time and was published in April 2003
- The finished genome was over 3 billion base pairs long but contained only about 25,000 genes
- The HGP discovered new data about non-coding DNA, suggesting that it plays an active role in the cell, and that it isn't just 'junk' DNA
- The sequence of the DNA is stored in databases available to anyone on the Internet
- At the same time as the HGP, teams of scientists set about the sequencing of the DNA of other organisms. This included the human gut bacterium, E. coli, the fruit fly and the mouse. Since then, more than 30 non-human genomes have been sequenced
Applications of the Human Genome Project
- Three key impacts of the HGP include:
- How many individual genes we have and how they work
- Locating the cause of genetic disorders
- Development of the new discipline of bioinformatics (the storage, manipulation and analysis of biological information via computer)
- The sequencing of the human genome has shown that all humans share the vast majority (99.9%) of their base sequences, but also that there are many SNPs that contribute to human diversity
- The information generated from the HGP has been used to tackle human health issues with the end goal of finding cures for diseases
- Scientists have noticed a correlation between changes in specific genes and the likelihood of developing certain inherited diseases
- Several genes within the human genome have been linked to increased risk of certain cancers
- There have also been specific genes linked to the development of Alzheimer's disease
- Ethical, legal and social issues are generated by the project
Genome Sequencing Techniques
NOS: Developments in scientific research follow improvements in technology; gene sequencers, essentially lasers and optical detectors, are used for the sequencing of genes
- In scientific research, critical developments often follow improvements in scientific apparatus
- For example, distant objects in Space often remain undiscovered until a telescope (or some other piece of equipment) powerful enough to detect them is developed
- The fact that scientific research is often held back by a lack of sufficiently powerful or precise apparatus is a problem that will continue into the future
- In some ways, this is very exciting, as it suggests that our scientific knowledge and understanding of the universe will continue to expand as new scientific techniques and technologies are developed
- Investigations such as the Human Genome Project are dependent on the use of powerful computers and improvements in technology to store and analyse vast quantities of data
- To sequence a genome:
- The entire genome is broken up into manageable pieces and then the fragments are separated so that they can be sequenced individually
- Single-stranded copies are made
- Nucleotides are each tagged with a differently coloured fluorescent marker, one for each base, adenine, cytosine, guanine and thymine
- Samples are separated according to length, by capillary electrophoresis machine. This procedure is very high resolution and distinguishes DNA fragments that differ in size by only a single nucleotide
- After separation, a laser beam makes the fluorescent markers fluoresce
- Then an optical detector linked to a computer deduces the base sequence from the sequence of colours detected
- This process highlights the use of a database to determine differences in the base sequence of a gene in two species
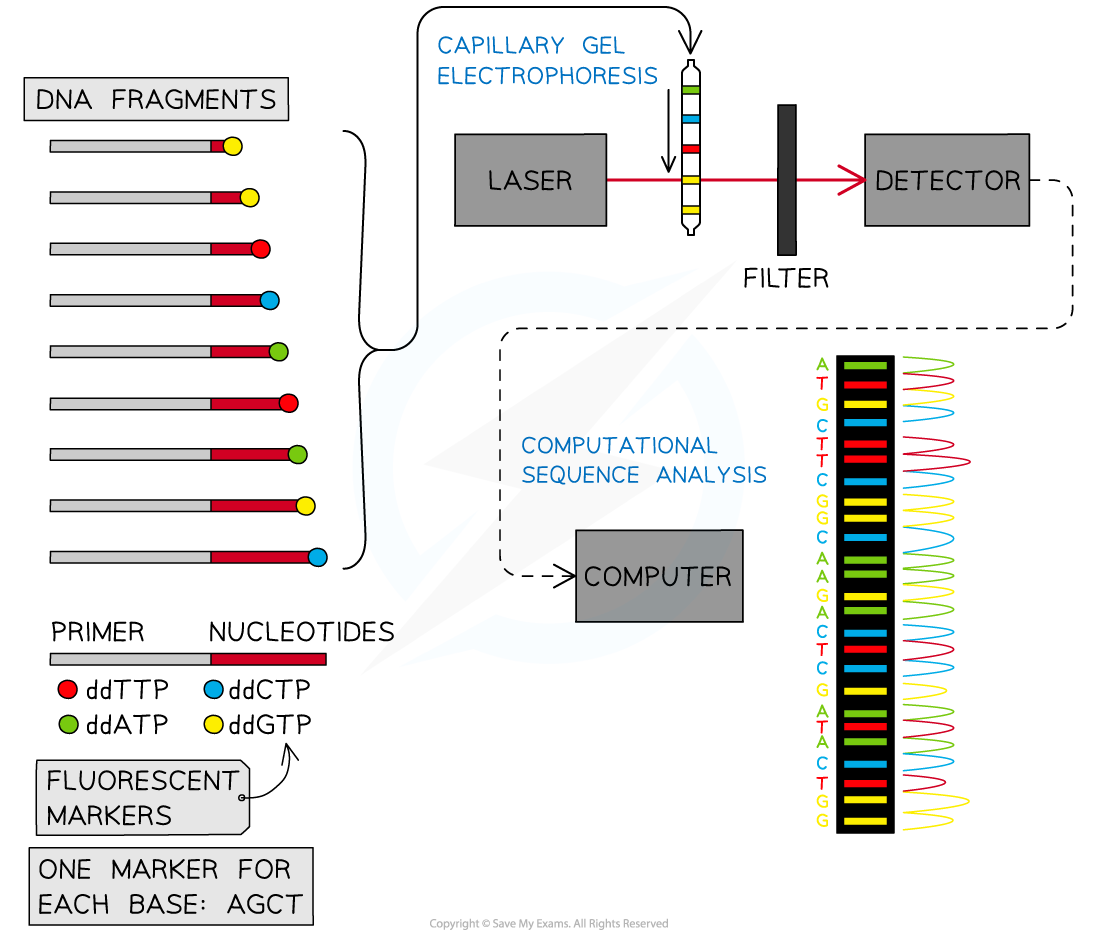
Gene sequencing is made possible by the use of optical laser technology, optical detectors, and computers
转载自savemyexams
早鸟钜惠!翰林2025暑期班课上线

最新发布
© 2025. All Rights Reserved. 沪ICP备2023009024号-1








