- 翰林提供学术活动、国际课程、科研项目一站式留学背景提升服务!
- 400 888 0080
IB DP Biology: HL复习笔记7.1.6 Skills: Nucleosomes & Molecular Visualisation Software
Skills: Nucleosomes & Molecular Visualisation Software
- Molecular visualisation software can be used to help understand molecular structures
- Macromolecules like protein, DNA, RNA and complex carbohydrates can be visualised as 3-D structures
- This allows researchers to analyse macromolecules and/or study interactions between them
- Primary sequence information can be related to structure and function
- This helps to relate how structure might relate to chemical or biological behaviour
- Macromolecules can be represented in many different ways including ball and stick atom models or simplified ribbon representations that show the protein backbone
- Most molecular visualisation software is freely available on the Internet or can be accessed through many bioinformatics repositories such as the Protein Data Bank (PDB)
Analysing the association between protein and DNA within a nucleosome
- Visit the Protein Data Bank PDB site and search for: 6T79 structure of human nucleosome (do not put the search term in quotes)
- Select the “3D view” to view the protein structure in mol*
- The 3-D structure of the nucleosome can be viewed
- The DNA double helix can be clearly seen surrounding the histone proteins
- Rotate or zoom into the image to visualise the different components
- The DNA can be seen to make two loops around the histone octamer core
- Look carefully - the tails of each histone protein can be seen projected from the nucleosome core
- These can be chemically modified to help regulate gene expression
- Try changing different settings in the viewer or select a different viewer such as JSmol
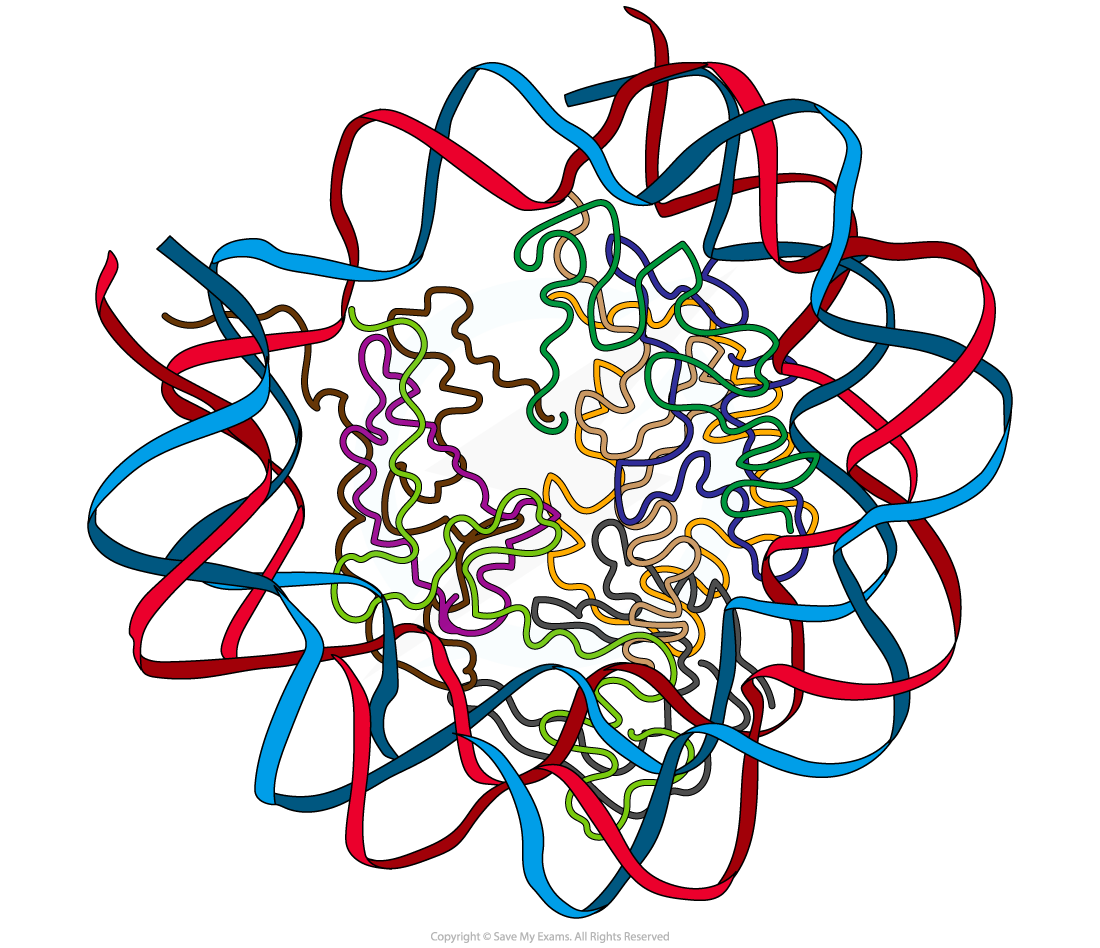
Structure of human nucleosome yeast tRNA showing the association between DNA (in 2 loops around the edge) and histones (central region)
转载自savemyexams

最新发布
© 2025. All Rights Reserved. 沪ICP备2023009024号-1









